MESH (Molecular Evaluation Science Hub)
Every week on Linkedin and other social media there is a new exciting generative model or molecular simulation or binding prediction tool released (click here to see just my stream). Exciting times!


It’s hard to keep up! New tools require testing and evaluation, and while we can work to automate a lot of the evaluation, at some point humans need to experiment with and test these tools in their own workflows.
Competitions: complex tools require social-technical collective evaluation
Academic and industry groups have used competitions to leverage our collective focus. They bring people together with both competitive fun, and collective learning. Some recent examples:
rotein designers from all around the world to designed a new binding protein against EGFR, a transmembrane protein that plays a critical role in cell growth, differentiation, and surviva
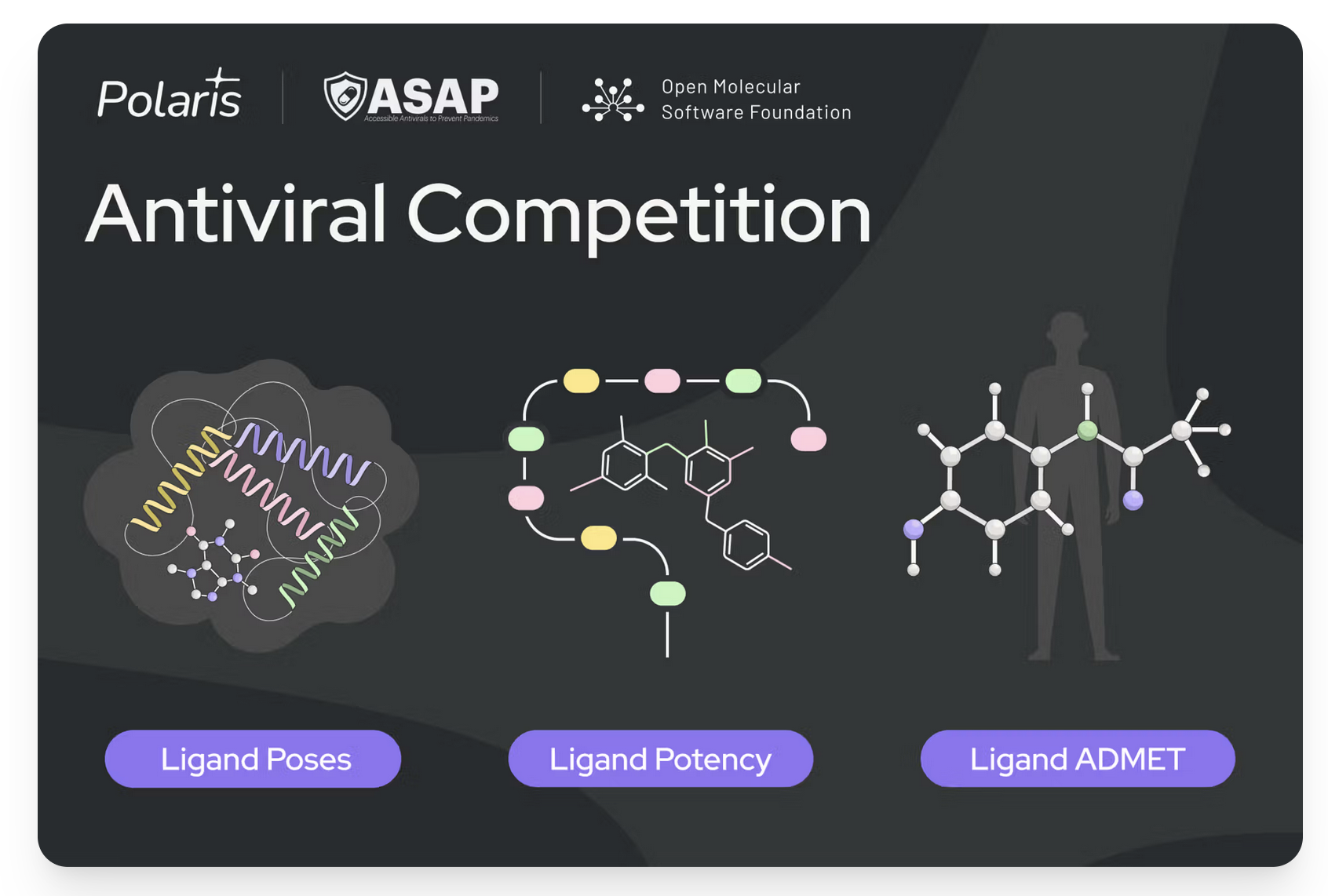
ASAP Discovery is an NIH-funded consortium leveraging open science for antiviral drug discovery, aiming for equitable and affordable global access to effective antivirals
Public competitions are an excellent way to learn from each other and channel resources. Bits to Binders was another one of these kinds new challenges/competitions, and I threw my hat in the ring!

Myself and 4 others joined this challenge to form a team that would generate a protein binder to a specific immune cell receptor, where a binder that would bind just right (not too much, not too little) would have the potential to treat some specific cancers.
Our team had access to GPU resources, the target, and a brand new generative AI model for protein design: RFDiffusion. None of us previously knew each other, and all come with different backgrounds and experience and technology
However, despite our enthusiasm, our team, and many like ours, encountered significant struggles. Internally within group, we struggled to effectively share workflows and results. The organizers of the competition had done a great job to gather sponsors and organizations who collectively provided a significant number of valuable GPU resources.
We could only utilize a very tiny fraction of available GPUs due to both mundane and complex technical and logistical challenges. There was simply a lack of infrastructure to support this social-technical phenomenon. We wanted a vibrant community, and the organizers did an absolutely fantastic job, but the outcome of the competition specifically in respect to sharing the methodologies was hampered by a lack of suitable infrastructure.
Put simply, there is no universal, straightforward way to share a computational workflow that requires visualization and interactivity (due to molecular structures) also connected to GPUs running AI models and simulations, running seamlessly on whatever GPU resources are available to you. While workflow systems like Nextflow and others can be a piece of the puzzle, they lack specific features around sharing and publishing due to the inherent technical tradeoffs.
This missing connection means that innovation remained isolated in the groups and labs of origin, regardless of the desire to share them.
What about the competition leaderboard examples you posted
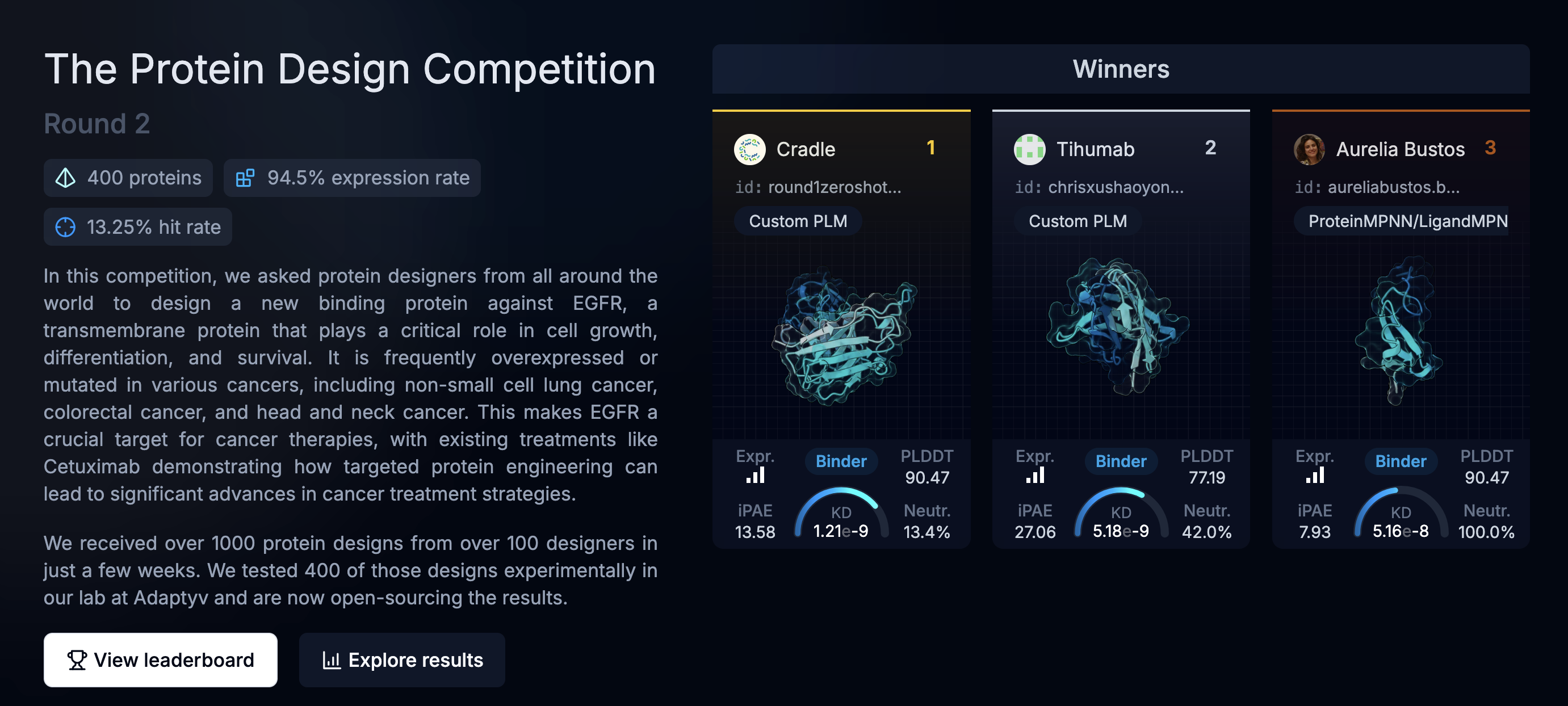
The Adaptyv Bio competition website looks fantastic, significant work and thought went into the design and infrastructure:
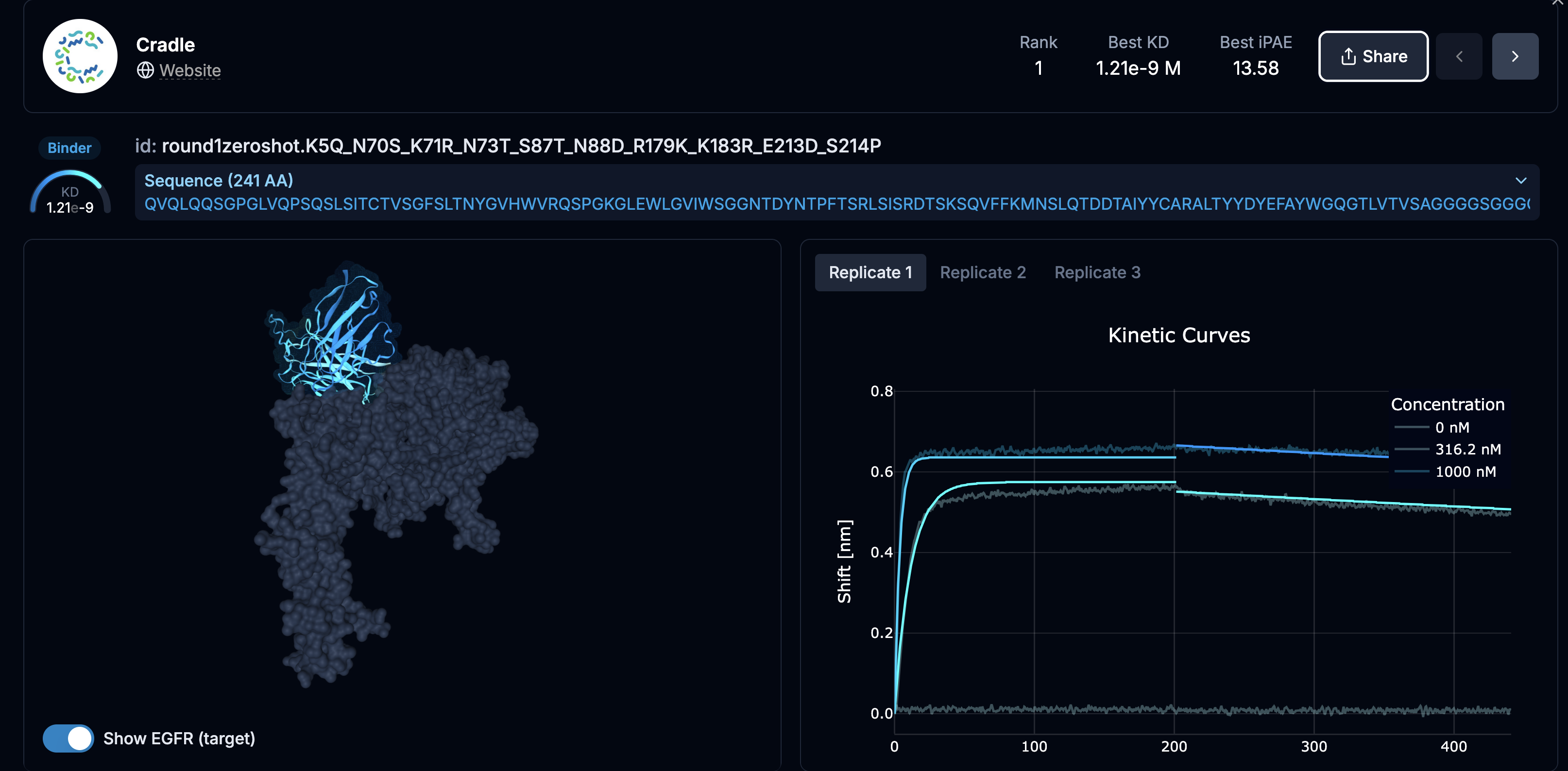
But digging into the well-designed pages and you find that the actual design process are not embedded, and often not available:

And that is part of the problem. Though well-designed, the competition site is a bespoke presentation, often not containing the methods, and results that are difficult to incorporate or evaluate systematically, and in the example above, there are no methods for you to copy and test yourself.
For the ASAP Discovery competition, for some entries there is code, but the </> code link is simply to a github repository. That means a lot of custom, bespoke work to understand, and possibly use those methods:
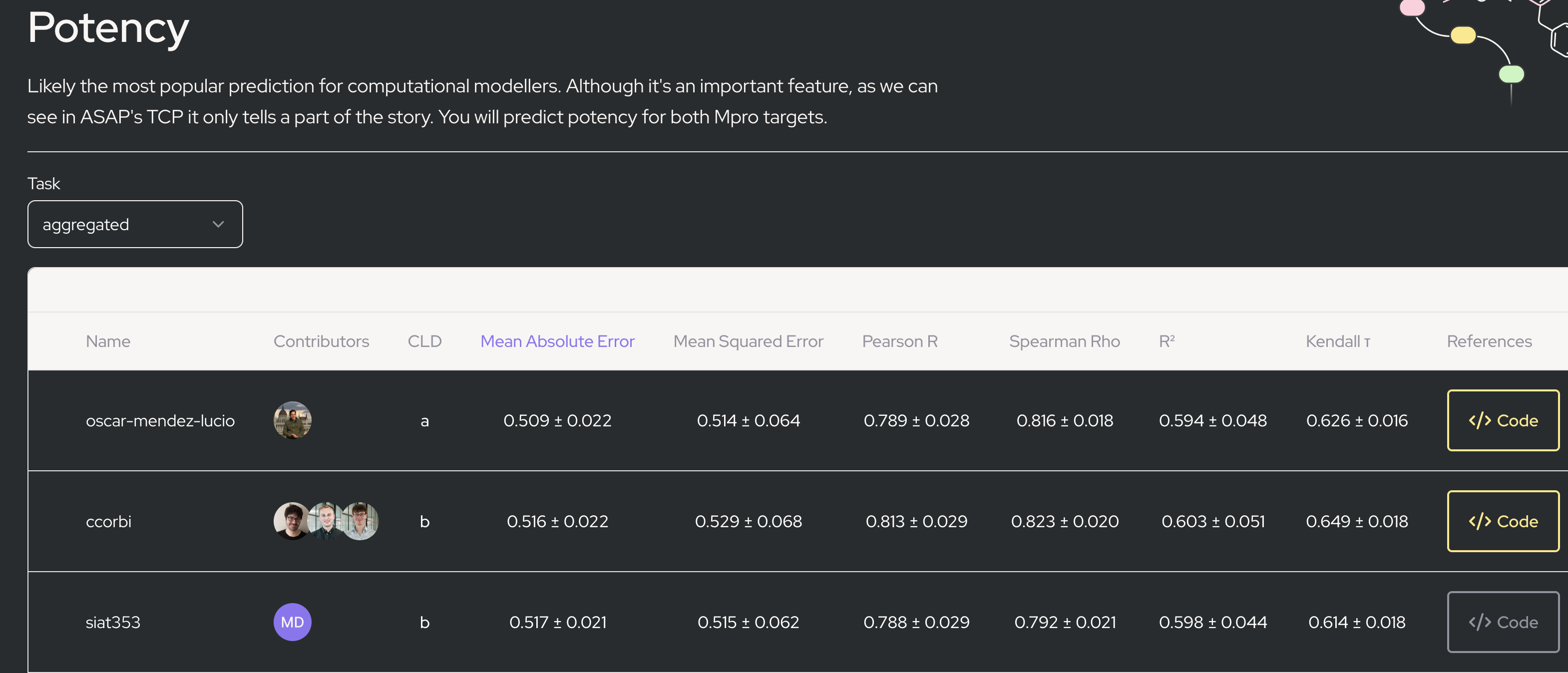
So in summary: you often cannot get methods and tools, and if you can the code is in a form that requires significant work just to run yourself. It’s too slow and limited, and this limits our ability to share and learn from each other, and it wastes a lot of repeated work, as everyone who wants to use a given tool has to do the same work (wasting significant effort) getting it working.
MESH (Molecular Evaluation Science Hub)
Recognizing these limitations, we propose a socio-technical initiative: MESH. Using some recent breakthroughs in publishing scientific workflows, we aim to harness the very best aspects of human collaboration: shared innovation and collective learning, and in this particular space, via leaderboards of workflows. These workflows are run and expressed in the browser, even if they are only headless workflows. The workflows can be shared, modified, and run entirely in the browser, even though there are backend containers running on some grid or cluster.
A central part of our vision is developing the collective set of open-source tools that express workflows directly in the browser: metapage workflows, combined with existing proven social structures for efficient idea exploration and refinement: public competitions and leaderboards that can rapidly and efficiently evaluate and spread to target communities novel and powerful molecular dynamics software tools, in a form they can immediately use.
The metapages platform and tools allow researchers to easily create persistent, durable, yet flexible leaderboards. These leaderboards aren't just static records; they are dynamic, adaptable workflows that researchers can easily manipulate, extend, and incorporate into their own workflows, in whole or in by parts. By leveraging a public compute grid, researchers can effortlessly connect their own or others computational resources, whether their own laptop, HPC clusters, or turn-key cloud solutions. This makes shared workflows independent of where they run (as long as you have or we provide the necessary compute resources), thus enhancing accessibility and reducing the barriers to participation and collaboration.
Example workflow in the browser
We have created an example dynamic leaderboard workflow, described in more detail here.

The leaderboard is fully dynamic. It can dynamically update all entries inputs, re-run the workflows directly from the browser, and automatically collates the results.
A leaderboard workflow of workflows!
All open-source, all re-usable, all running without any installation steps
This initiative is in the house of The Open Molecular Software Foundation whose mission statement is a succinct superset of ours. Our particular alignment is especially about accelerating progress. We envision empowering researchers of all technical backgrounds to seamlessly integrate innovations in molecular dynamics AI models developed by one group into the workflows of another, and they will be able to do this because they will be able to rely on tools that are more reliable, as the tools will have been (and can continue to be evaluated by a community. This democratization of AI tools and knowledge will unlock previously constrained collaboration potential, fueling advancements and discoveries across the molecular biology and broader scientific community.
We will follow up with a post about the technical differences between metapage workflows and other comparable stacks such as Jupyter Notebooks and workflow systems such as Nextflow.
References
Competitions with dashboards
https://foundry.adaptyvbio.com/competition
https://beta.adaptyvbio.com/benchbb
https://polarishub.io/blog/antiviral-competition
https://paperswithcode.com/method/clip

