A new way to publish interactive data and visualizations
Deciding where how how to publish your data intensive scientific workflows can be difficult, with no clear solution.
The most common solution is to publish a jupyter notebook somewhere. There are a number of platforms that
- Take a jupyter notebook
- Run it, get the outputs
- Convert the notebook and outputs into a static web page
For example, jupyterbook (using the myst-nb extension) compiles a jupyter notebook, with text and images, into a page for publishing. If you want to run the notebook yourself, you might be able to via binder or jupyterhub.
So, it looks like this is a solved problem? Actually, there are many limitations with these kinds of solutions, and they stem from starting with something not built for the web, and bolting on pieces to make it web-friendly. It works, but it ends up being complicated, and starts to show limitations.
Problems and limitations
Problem: the content is static
The above platforms generate the content for a fixed, static website. There are some options to run the notebook on binder or jupyterhub, but those platforms are ephemeral, and greatly limit the kind of compute instances the notebooks run on. Any data generated is lost unless specific ad-hoc steps are taken to keep generated outputs. What if I didn’t want to run the notebook on those platforms, for lots of reasons (not enough compute, storage, persistence, security), then I don’t have other options.
While some notebook visualizations are dynamic in some sense, as you can interact with e.g. javascript plots, it becomes complicated or limited if you want to perform computation in the final published notebook.
Limitation: you cannot easily take all or a part of the notebook for your own workflows
Jupyter notebooks run on a single kernel, with all the visualization libraries built into the monolith kernel.
If I wanted to swap out one visualization library for another (e.g. matplotlib for plotly.js) it’s quite a bit of coding.
There’s no ability for users to publish components, and re-use those components in different notebooks. Each notebook is a monolith, unless you publish a separate library, which comes with it’s own friction and cost.
You don’t just publish and share code and visualization snippets for others. Instead, you have to manually copy and paste and finagle any code from one notebook to another. And everyone has this same barrier.
There is considerable friction in sharing and re-using notebooks and parts of notebooks.
Limitation: a single kernel, not always containerized
Jupyter notebooks define their kernels, but without much automation to build those kernels. Take a look at this example of a recent published model for predicting structure distributions of a protein: each step requires the user input, and data is stored in Google Drive only. Interaction is via the notebook interface, so integrating the code into your own workflow is a manual process that every single person must repeat, if they want to use the model in their own workflows.
I would like to not have to ever manually install anything. Workflows should contain all instructions for how to build their required computation environments, and those instructions should be automated by default.
Limitation: you cannot easily collaborate real time
The platforms for sharing notebooks are meant as publishing as a final step. Making it easier to publish is a good thing, but publishing is really only a step in a sequence: you might want to publish so that others might be able to use your work. This process might go back and forth, but since publishing is treated as a final step, the collaborative aspect, which is often the bulk of the work, is neglected. The final published product is not easily integrated into others workflows without significant human labour.
Putting a notebook in a git repository is helpful, but not sufficient.
Problem: the complexity of the layers introduces more places for bugs and failures
Because the notebooks may not be properly containerized, it’s more likely to get build errors. The public platforms to host jupyter notebooks are by necessity limited, and it’s some work to run them yourself, so the surface area for bugs and incompatibilities is large.
Where to publish your code/data/visualization?
So that other researchers can actually use them? Publishing your work doesn’t automatically mean it can be found. There is no one place to search for code and data and workflows, so finding code snippets or visualization relevant for your problems is ad-hoc and a manual process.
Metapages: publish and embed complete scientific workflows, when you want, where you want, how you want
Metapages are web-first scientific workflows.
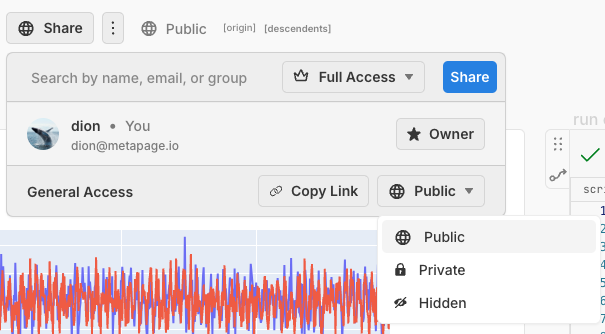
Publish in two clicks
You don’t need a complicated separate build/process/embed publish step required of e.g. jupyterbook. You just click Share → Public and your workflow is immediately published for everyone:
Embed anywhere
If you want to embed the workflow into your own notes or page or website or notion or anywhere, simply Share → Embed and paste the link or HTML snippet where you want. It can contain a link for navigating to the original
Here is an interactive workflow embedded. You can access (then copy and edit) the entire workflow with the menu button on the left, without any installation steps:
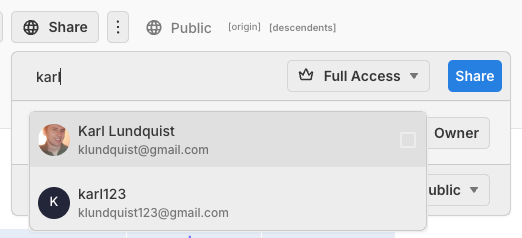
Permissions: collaborate with anyone
In a few seconds, grant permissions to your collaborators, they can now read or edit your workflow, upload/download data, and real-time work with you
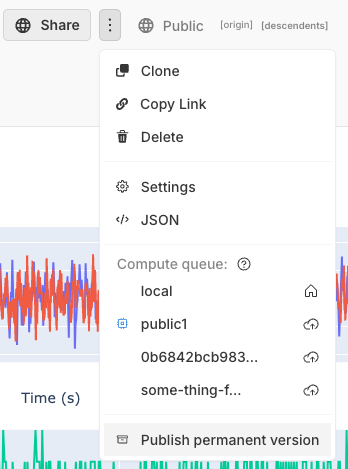
Publish permanent versions forever
This feature is in beta
When you want a permanent copy for publishing or archiving, simply create an archive, and we will store it forever
This will generate a permanent URL, with a sha256 hash of the workflow, similar to how git creates a hash of each commit. This URL will work, even if the underlying workflow is unpublished or deleted.
